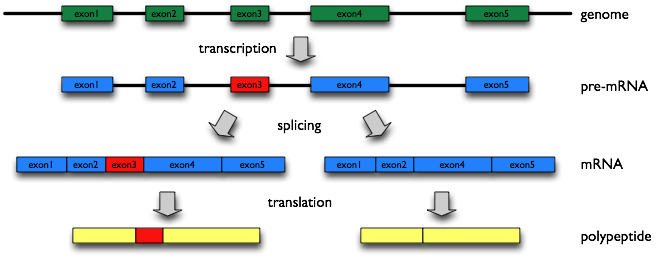
Alternative splicing (AS) is a molecular mechanism that produces multiple proteins from a single gene, and is thought to increase the number of proteins extensively over the number of genes in a genome, which could be required to generate complexity of higher eukaryote including human. However, what roles most of AS variants play are not known.
This database, AS-ALPS (Alternative Splicing-induced ALteration of Protein Structure), is aimed at providing useful information to analyze effect of AS on protein interaction and network through alteration of protein structure.
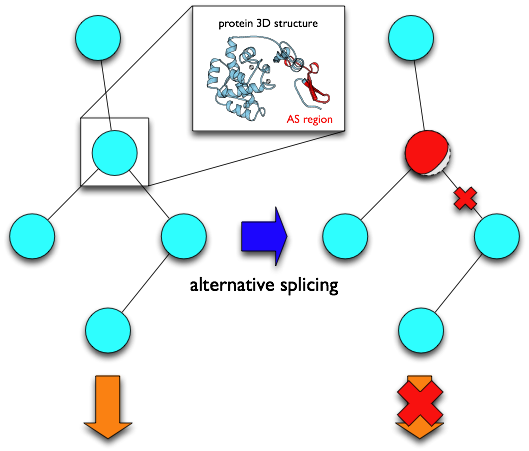
In AS-ALPS, regions of amino acid sequences changed by AS ("AS regions") which we detected with transcript sequences in RefSeq, Ensembl, etc., are linked to information extracted from PDB about residues forming hydrophobic cores and inter-molecular interaction sites. This makes it possible to directly infer whether protein structure and/or interaction are affected by each AS event. In addition, AS-ALPS provides links to a protein network database KEGG, making it easy to know which network and which node in the network can be influenced by AS.
AS Data of ver. 1.8.4 can be downloaded here.
| Species | AS regions | AS variants (mRNAs) | AS isoforms (proteins) |
|---|---|---|---|
| H.sapiens | hs_AS_regions.txt.gz | hs_AS_variants.txt.gz | hs_AS_isoforms.txt.gz |
| M.musculus | mm_AS_regions.txt.gz | mm_AS_variants.txt.gz | mm_AS_isoforms.txt.gz |
| D.melanogaster | dm_AS_regions.txt.gz | dm_AS_variants.txt.gz | dm_AS_isoforms.txt.gz |
| C.elegans | ce_AS_regions.txt.gz | ce_AS_variants.txt.gz | ce_AS_isoforms.txt.gz |
| A.thaliana | at_AS_regions.txt.gz | at_AS_variants.txt.gz | at_AS_isoforms.txt.gz |
| O.sativa | os_AS_regions.txt.gz | os_AS_variants.txt.gz | os_AS_isoforms.txt.gz |
| Swiss-Prot | sp_AS_regions.txt.gz | - | sp_AS_isoforms.txt.gz |
If you use AS-ALPS in your work, we request you to cite the following paper and the URL (http://as-alps.nagahama-i-bio.ac.jp/).
Methodologies to detect AS region and annotate AS regions are described in the following paper.
This database was supported by grants of the Genome Network Project, Grants-in-Aid for Scientific Research (B) and Grant-in-Aid for Young Scientists (B) from Ministry of Education, Culture, Sports, Science and Technology of Japan.